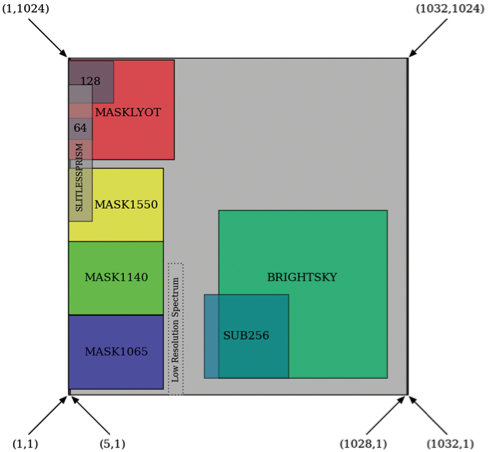
MIRI Detector Subarrays
MIRI imaging, coronagraphic imaging, and low-resolution spectroscopy utilize a pre-defined set of 9 subarrays for different observing strategies, each with its own advantages and recommended uses.
On this page
See Also: Understanding Exposure Times
MIRI’s detector arrays have the ability to read out partial frames through the manipulation of clocking patterns. Subarray readouts reduce the frame time to less than the nominal 2.775 s per full frame. The portion of the full array that forms the subarray is read out and stored while the remaining parts of the array are reset.
Nine different subarrays are available for imaging, coronagraphic imaging, and low-resolution spectroscopy (LRS): 4 separate subarrays for coronagraphs, one for high background, 3 for bright objects, and one slitless LRS spectrum. No subarrays are available for the medium-resolution spectrometer (MRS).
Subarray readout
See also: MIRI Detector Readout Overview, MIRI Detector Readout FASTR1, MIRI Detector Readout SLOWR1
Subarray coordinates cannot be randomly accessed so the row and column shift registers must step from the origin (1,1) to the starting subarray corner before proceeding. In other words, at the beginning of each frame read, the first two rows of the full array are accessed briefly and reset, then the 2nd pair, etc., until the subarray is reached.
In the first row of the subarray, pixels on the left that are not part of the subarray are clocked through (and not digitized), then the pixels that are part of the subarray are read. Pixels to the right of the subarray are ignored by resetting the column shift register to 0 immediately after the last subarray pixel. This pattern is repeated through all the rows contained within the subarray. The rows after the subarray are stepped through quickly and reset as were the rows before the subarray. It follows that subarrays are slower the farther they are from the left edge of the array.
This readout scheme drove the orientation of the imager array, since it is advantageous to have the fastest subarrays located within the coronagraph. It also means that it takes exactly the same amount of time to read a 256 × 256 subarray starting at (257,257) as it does to read a 512 × 256 subarray starting at (1,257); the only difference is the amount of data passed to the solid state recorders and the ground. Other issues that determined the subarray location included: the ability to utilize reference pixels, minimizing dead time from the clocking to access the first pixel, bad pixels on the array, and the best imaging location for the optics.
For more efficient subarray operation, a “burst mode” clocks through the left columns at 5 times the normal speed. In the first row of the region of interest (ROI), pixels on the left that are not part of the ROI are clocked through (and not digitized), after which the pixels that are part of the ROI are read. Pixels to the right of the ROI are ignored by resetting the column shift register to zero (recall the special shift register definition) immediately after the last ROI pixel. This pattern is repeated through all the rows contained within the ROI. The rows after the ROI are stepped through quickly and reset as are the rows before the ROI.
Imaging
See also: MIRI Imaging, MIRI Imaging Template Parameters
Words in bold are GUI menus/
panels or data software packages;
bold italics are buttons in GUI
tools or package parameters.
Table 1. MIRI imaging subarray characteristics
| Subarray | Size in pixels (rows × columns) | First row corner | First column corner | Usable size | Group time† |
|---|---|---|---|---|---|
FULL | 1024 × 1032 | 1 | 1 | 74" × 113" | 2.775 s |
| BRIGHTSKY | 512 × 512 | 51 | 457 | 56.3" × 56.3" | 0.865 s |
| SUB256 | 256 × 256 | 51 | 413 | 28.2" × 28.2" | 0.300 s |
| SUB128 | 128 × 136 | 891 | 1 | 14.1" × 14.1" | 0.119 s |
| SUB64 | 64 × 72 | 779 | 1 | 7" × 7" | 0.085 s |
† Group times are calculated for FASTR1 mode only. SLOWR1 mode readout will only be available for the FULL array.
Coronagraphic imaging
See also: MIRI Coronagraphic Imaging, MIRI Coronagraphic Imaging Template Parameters
In coronagraphic imaging, each subarray is tied to a specific filter. The subarray choice is therefore hidden from the user in the JWST Astronomer's Proposal Tool (APT).
Table 2. MIRI coronagraphic imaging subarray characteristics
| Subarray | Filter | Size in pixels | First row corner | First column corner | Size‡ | Group time† |
|---|---|---|---|---|---|---|
| MASK1065 | F1065C | 224 × 288 | 21 | 1 | 24" × 24" | 0.240 s |
| MASK1140 | F1140C | 224 × 288 | 243 | 1 | 24" × 24" | 0.240 s |
| MASK1550 | F1550C | 224 × 288 | 463 | 1 | 24" × 24" | 0.240 s |
| MASKLYOT | F2300C | 304 × 320 | 719 | 1 | 30" × 30" | 0.324 s |
‡ This angular size is the illuminated FOV. The subarray size listed in pixels is the true size of the array (in pixels), which defines the exposure time per frame.
† Frame times are calculated for FASTR1 mode only. SLOWR1 mode readout is only available for the FULL array.
Low-resolution spectroscopy
See also: MIRI Low Resolution Spectroscopy, MIRI Low Resolution Spectroscopy Template Parameters
The size of the slitless prism subarray is determined by the number of pixels needed to cover the 5–14 μm low-resolution spectrometer spectrum in the dispersion direction and provide adequate sky observations for background subtraction in the spatial direction.
Table 3. MIRI low-resolution spectroscopy imaging subarray characteristics
| Subarray | Size in pixels | First row corner | First column corner | Group time† | Notes |
|---|---|---|---|---|---|
| FULL | 1024 × 1032 | 1 | 1 | 2.775 s | Slit spectrum |
| SLITLESSPRISM | 416 × 72 | 529 | 1 | 0.159 s | Slitless spectrum |
† Frame times are calculated for FASTR1 mode only. SLOWR1 mode readout is only available for the FULL array.
References
Ressler, M. E. et al. 2015, PASP, 127, 675
The Mid-Infrared Instrument for the James Webb Space Telescope, VIII: The MIRI Focal Plane System